Edit Content
Trending




A new study published in Biochemistry sheds light on how bacteria regulate their genes, challenging long-held assumptions about protein behavior. The research compares how two bacterial species—Escherichia coli and Mycobacterium tuberculosis—use a signaling molecule called cyclic AMP (cAMP) to control important cellular functions.
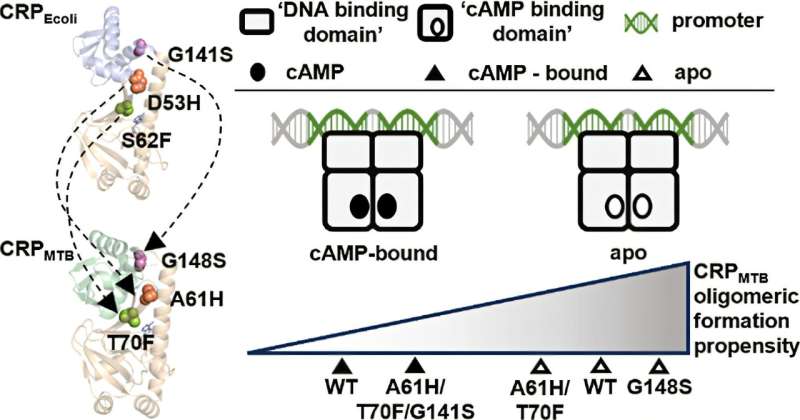
The team provides key insights into how bacterial cAMP receptor proteins (CRPs) respond differently to the ubiquitous signaling molecule, cyclic AMP (cAMP). By comparing the allosteric regulation of Escherichia coli CRP (CRPEcoli) and Mycobacterium tuberculosis CRP (CRPMTB), researchers challenge the assumption that structural similarity predicts functional behavior in allosteric proteins.
This study introduced structurally homologous mutations from previously identified allosteric hotspots in CRPEcoli into CRPMTB to investigate their effects on protein solution structure, stability, and function. The results reveal that despite their structural resemblance, these two proteins from distantly related bacteria do not share identical allosteric behaviors or hotspots in response to molecular signals. These findings underscore the complexity of allosteric regulation and suggest that species-specific mechanisms drive bacterial gene regulation.
Beyond advancing fundamental knowledge of bacterial signaling, this research has potential implications for drug development. Understanding the unique allosteric properties of CRPMTB may inform therapeutic strategies that target transcriptional regulators in M. tuberculosis, a critical factor in bacterial adaptation and pathogenesis.
The research, titled “Identifying Allosteric Hotspots in Mycobacterium tuberculosis cAMP Receptor Protein through Structural Homology,” was led by Dr. Rodrigo Maillard at Georgetown University.
Future research will explore how evolutionary differences shape allosteric signaling pathways across bacterial species. Understanding the unique regulatory mechanisms of CRP in M. tuberculosis compared to E. coli may provide insights into bacterial adaptation and gene regulation. Additionally, identifying novel regulatory elements within CRP-mediated pathways could inform antimicrobial strategies aimed at selectively disrupting essential bacterial signaling networks, potentially leading to more targeted therapeutic approaches.
More information:
Stephen P. Dokas et al, Identifying Allosteric Hotspots in Mycobacterium tuberculosis cAMP Receptor Protein through Structural Homology, Biochemistry (2025). DOI: 10.1021/acs.biochem.4c00723
Provided by
Georgetown University Medical Center
Citation:
Bacterial species study challenges assumption that structural similarity predicts protein behavior (2025, February 7)
retrieved 8 February 2025
from https://phys.org/news/2025-02-bacterial-species-assumption-similarity-protein.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
©2024. Livebuzznews. All Rights Reserved.